
Cut a word out of this sentence and do anything you want with it. Insert it somewhere else in this article. Throw it away. If you’re feeling ambitious, make a few copies of it and disperse them across the article, or scramble the letters before putting the word back in place.
If you were to make these alterations at random, most of the resulting text would be nonsense. Making a grammatically correct change would be unlikely, and improving the existing article would be almost impossible. Yet at the level of DNA, which is written in a chemical language with letters called bases, nature depends on these essentially random changes to make adaptations to organisms over time.
These modifications, called mutations, are the cornerstone of evolution. If a mutation increases the odds that its owner will reproduce, it will be propagated when that organism’s genetic material is transmitted to its offspring. As they are passed down to each successive generation, mutations ensure that life can adapt and expand to fill new niches.
But mutations are mistakes—errors made when DNA is copied or consequences of chance chemical reactions. Mutations don’t occur in response to environmental cues, making an organism’s offspring fitter by default. Instead, mutations may strike any part of an organism’s DNA, and many mutations are neutral or even harmful. A fortuitous mutation that improves a single gene is rare by comparison, and many adaptations require mutations in multiple genes.
Put this way, it seems outlandish that mutations are responsible for the complexity and variety of life on Earth, yet they are. Life has devised shortcuts that increase the impact and usefulness of individual mutations, and the iterative nature of evolution amplifies small genetic changes over time. Each mechanism is molecular, occurring at the microscopic scale of DNA. But the impacts are far-reaching, responsible for the genetic differences between one person and another, between a bacterium and a fly.
In search of the molecular basis of evolution, scientists are investigating the largest genetic leaps, from the origins of multicellularity to the rapid and flexible adaptation of animal body plans. Others are digging even further into the history of life, to the humble bacteria that made some of the earliest contributions to molecular evolution—billions of years in the past.
Digging for molecular fossils
Shion Lim, a graduate student in Professor Susan Marqusee’s lab in the Department of Molecular and Cell Biology at UC Berkeley, is investigating the ways mutations drive the evolution of proteins, an important type of molecule in living things. Proteins, which are made of smaller molecules called amino acids linked together in specific sequences, come in many varieties. The instructions for how to make each type of protein are written in an individual segment of DNA called a gene, so a mutation in a certain gene can cause a change in the corresponding protein.
“The amino acid sequence of the protein dictates the properties of the protein,” Lim says. Understanding how the sequence changes over time provides clues about how proteins gain new functions. By studying old versions of modern proteins, she hopes to understand the way mutations accumulated over generations. But this molecular fossil hunt is complicated by the fact that proteins degrade rapidly compared to the pace of evolution, so researchers have to infer what ancient proteins were like based solely on proteins that exist today.
To unfold the mystery, Lim uses a technique known as ancestral sequence reconstruction, which involves comparing amino acid sequences of proteins in existing organisms that are distantly related. Based on information about the evolutionary relationships among many kinds of bacteria and the probabilities that various types of mutations will occur, Lim determines the most likely sequence of the ancestral protein. Once the sequence is known, Lim says, it’s even possible to create the ancient protein in the lab and compare its properties to those of its descendants.
 Climbing down a tree from its branches: In ancestral sequence reconstruction, researchers compare modern proteins (blue, green and yellow shapes on branches) to predict the properties of an ancient protein (blue shape on trunk). The ancient protein can then be resurrected in the lab and studied. Image Credit: Ashley Truxal
Climbing down a tree from its branches: In ancestral sequence reconstruction, researchers compare modern proteins (blue, green and yellow shapes on branches) to predict the properties of an ancient protein (blue shape on trunk). The ancient protein can then be resurrected in the lab and studied. Image Credit: Ashley Truxal
By studying ancestral proteins at different branch points on the evolutionary tree, Lim hopes to find out how incremental changes to a protein’s sequence change its properties, potentially leading to adaptations. Understanding how proteins help organisms adapt can also provide clues about prehistoric environments. For instance, Lim says, many researchers have hypothesized that Earth was very hot when life was first being established over three billion years ago—but her research on bacterial proteins suggests otherwise.
To get an idea of the environment in which ancient organisms lived, Lim applied ancestral sequence reconstruction to a protein called RNase H from two modern bacterial species that live in very different environments. One species, called Thermus thermophilus, thrives at a scalding 149°F. In contrast, the more familiar Escherichia coli grows best at around 98.6°F, the body temperature of its human host. Unsurprisingly, when she compared the bacterial proteins’ ability to withstand heat, the T. thermophilus version of the protein fared better.
If Earth was extremely hot 3.2 billion years ago, when the T. thermophilus and E. coli lineages split, the ancestral version of the protein should be as stable at high temperatures as the T. thermophilus protein. Instead, Lim says, the reconstructed protein exhibited heat tolerance halfway between that of the heat-loving and more moderate bacteria, making a warm—but not sweltering—ancient Earth more likely. And while more work is needed to further characterize this environment, Lim says that some other groups have reported similar findings.
Regardless of the exact features of the ancient world, the differences between the lab-resurrected protein and its modern descendants illustrate the role of mutations in protein evolution. Both of the bacterial lineages slowly accumulated mutations that modified an existing protein, maintaining the protein’s original function while making it better adapted to its environment.
“I think that is really the basis of molecular evolution—that mutations affect what proteins do,” Lim says. “That ultimately changes the fitness of the protein in the cell, which in turn contributes to how fit the organism is to survive and generate offspring.” Such changes are critical to the success of a species. But explaining major evolutionary breakthroughs, such as the development of new proteins with unique functions, is more complex.
Chemical copies get sloppy
Concocting a new protein completely from scratch requires a series of random mutations in nonessential DNA somewhere in the genome. Although possible, this approach is slow. A faster method is to mutate an existing gene in a way that modifies its corresponding protein’s function, perhaps by subtly changing how it works or altering its location in the cell. Although it’s quicker, this method comes at a price: the function of the original protein is lost.
But nature has invented a workaround. Another type of mutation, called a duplication, can generate additional copies of an existing gene. Mutations can then accumulate in the redundant gene without overwriting the original. This leaves the copied gene free to explore different, potentially useful sequences, which can ultimately result in a novel protein. By reusing beneficial genes, nature is able to capitalize on prior successes, reducing the barrier to evolving new proteins and, in turn, to developing adaptations. And this, says Lim, makes gene duplication one of the driving forces behind protein evolution.
 Changing a few base pairs could decide the fate of this fish - be eaten by a shark or swim on to reproduce. Credit: Ashley Truxal
Changing a few base pairs could decide the fate of this fish - be eaten by a shark or swim on to reproduce. Credit: Ashley Truxal
Modifying individual genes one by one, little by little, has the potential to generate a diverse array of proteins with the variety of functions needed to sustain life—given sufficient time. But in nature, time is limited. Climates fluctuate, new competitors for resources appear, and the strategies that worked two generations ago might not work today. To make it this far, nature has had to create additional techniques to generate useful new combinations of genes.
Bacteria, for instance, can transfer entire genes or groups of genes by swapping circular DNA molecules called plasmids. Unfortunately for us, some of the most popular plasmids shared among bacteria include genes that confer antibiotic resistance, which makes pathogenic bacteria able to survive treatment by various classes of drugs.
Worse yet, bacteria reproduce rapidly, increasing the rate at which new antibiotic resistance genes materialize. The rise of antibiotic resistance is an evolutionary process we can actually witness in action, but one of the secrets to combating it might lie deep in bacteria’s evolutionary history. According to Lim, a very long-term goal of ancestral sequence reconstruction is to understand how antibiotic resistance genes evolve, allowing us to stay one step ahead of bacteria.
“In evolution, it’s an arms race between antibiotics and pathogens,” Lim says. “Can we actually predict what the next change would be?” Although it will likely be a long time before we can foresee a pathogen’s next moves, the promise of these future applications combined with established drug discovery techniques may mean our battle with pathogenic bacteria is not a hopeless cause.
Building cellular societies
In the world of bacteria, it’s a free-for-all: every cell for itself. Temporary alliances may exist, but true cooperation is rare. Getting groups of cells to work together, one of the few things most bacteria don’t seem to have mastered, is a key contributor to the evolutionary success of myriad life forms.
When single cells join forces to form a multicellular organism, they can become specialized. Different cells may take on varying roles, and by working together, cells can share resources and protect each other. Although they typically take much longer to multiply than single-celled creatures, multicellular organisms are equipped with an array of tools that make them more likely to survive, allowing them to fill unique niches. But while multicellularity’s advantages are clear, the way it emerged in the first place is murkier.
In humans, for example, interactions among cells depend on hundreds of genes, many of which weren’t even in the genomes of the first multicellular animals. Understanding how the last common ancestor shared by all animals first made the jump to multicellularity would fill in a major transition in our lineage’s evolutionary history. According to Tera Levin, a former UC Berkeley graduate student who is now a postdoctoral fellow in Professor Harmit Malik’s lab at the Fred Hutchinson Cancer Research Center, multicellularity “basically served as the foundation for everything that later evolved in animals.”
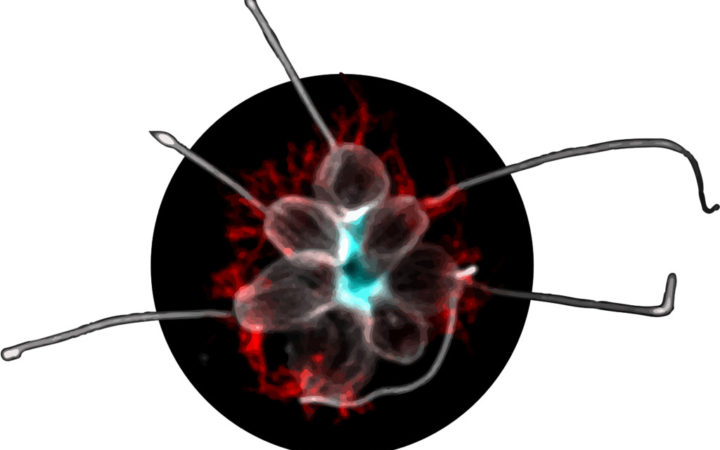 A microscope image shows the multicellular form of a choanoflagellate composed of several individual cells. Rosetteless, a protein that helps the cells stick together, is stained blue. Early animals may have used a similar method to develop multicellularity. Credit: Tera Levin
A microscope image shows the multicellular form of a choanoflagellate composed of several individual cells. Rosetteless, a protein that helps the cells stick together, is stained blue. Early animals may have used a similar method to develop multicellularity. Credit: Tera Levin
It isn’t trivial to dissect the molecular mechanisms behind the initial cooperation among cells. Just as ancestral proteins degrade, so do ancient organisms—and their genetic information vanishes along with them. Although animals’ earliest ancestor is long gone, researchers in Professor Nicole King’s lab in the Department of Molecular and Cell Biology at UC Berkeley have developed a clever approach to deducing what it may have been like. In this lab, where Levin conducted her PhD research, scientists are studying the closest living relatives of modern animals: the choanoflagellates.
Choanoflagellates possess a rudimentary form of multicellularity—they are single-celled creatures that can be coaxed to become multicellular by an environmental trigger. Given the right conditions, when a choanoflagellate undergoes cell division, the resulting cells stay in proximity to one another. Over repeated divisions, this process results in a cooperative globule called a rosette, which may consist of dozens of cells. If the rosettes are assembled via a prototypical version of the mechanism modern animals use to maintain a cohesive whole, the similarities could be used to determine which mutations allowed early animals to develop multicellularity.
In search of the genes responsible for rosette formation, Levin and fellow researchers in the King lab hunted for choanoflagellates that couldn’t be triggered to cooperate. They divided a group of genetically identical choanoflagellates into separate culture containers and bombarded them with radiation to induce mutations, hoping that one mutation would incapacitate a gene essential for rosette formation. Next, they visually screened an astonishing 15,344 separate cultures for cells that are incapable of forming rosettes but don’t have any other obvious defects. One of their mutants met these criteria, and further genetic analysis revealed that the impairment in rosette formation resulted from mutations that disabled a single gene.
The gene encodes a protein the researchers named Rosetteless, which Levin and colleagues discovered is located right in the center of rosettes, a region filled with molecules that hold the cells together. Asked what this implies about the mechanism of rosette formation, Levin comments: “I suspect that Rosetteless is a component that serves to connect and stabilize [the molecules in the center of rosettes], which is part of what gives structural integrity to the rosette colony.”
Since Rosetteless is a member of a group of proteins known to help animal cells stick together, early animals may have evolved to be multicellular using a similar method. Today’s animals have many proteins of the same class that serve additional roles, illustrating the use of gene duplication to create molecular diversity. So although multicellularity in modern animals relies on hundreds of genes, the critical first step may have required just one.
Enhanced evolution
While mutations within genes are responsible for a large portion of evolutionary change, another source of variation lurks outside the coding regions of the genome. In the genome, as in the written word, context counts. To determine whether a gene should be activated to produce the protein it encodes, the cell relies on the surrounding information—the noncoding DNA.
An organism’s genome isn’t solely composed of freestanding genes. Beyond the limits of genes lie DNA sequences called enhancers, which affect how and when a gene will be read, the first step toward making the protein the gene encodes. Specialized proteins called transcription factors scour the genome looking for specific enhancers, and when they find the right DNA sequence, they latch on. After this binding event, any gene controlled by the enhancer is read much more frequently, often by several orders of magnitude.
Because they allow an extra layer of control over genes, enhancers can be genetic boons for any creature. But the most ingenious use of enhancers to accelerate the pace of evolution was invented by physically sophisticated multicellular life forms. They use enhancers to regulate development, a process that simpler organisms lack. Building an animal’s leg or liver, for example, requires much more extensive genetic instructions than any bacterium could muster.
In animals, transcription factors often act on many enhancer sequences at once, effectively managing dozens or even hundreds of genes at a time. Many of these transcription factors regulate sets of genes used in development, such as those that instruct a cell whether to act more like a skin cell or a muscle cell, or whether a given section of the body will grow legs or arms.
In Professor Nipam Patel’s lab in the Department of Integrative Biology at UC Berkeley, graduate student Erin Jarvis is investigating the relationship between one type of these transcription factors, the Hox proteins, and limb development in the sand flea Parhyale hawaiensis. In a study led by Julia Serano, a former research specialist in the Patel lab, Jarvis and coworkers discovered that during development, each segment of this tiny crustacean contains a specific set of Hox proteins. Intriguingly, they found that the cocktail of Hox proteins in a given segment has a direct correlation to the type of appendage found in that segment.
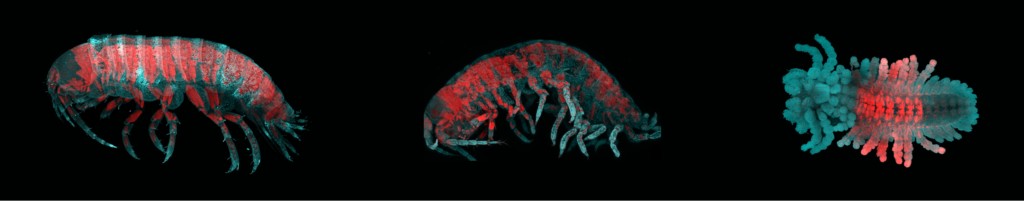 Left: A normal sand flea. Middle and Right: Sand fleas missing one of the Hox proteins (Abdominal-B), leading to loss of identity of abdominal body segments. Swimming legs and uropods are replaced with walking legs. Image credit: Erin Jarvis
Left: A normal sand flea. Middle and Right: Sand fleas missing one of the Hox proteins (Abdominal-B), leading to loss of identity of abdominal body segments. Swimming legs and uropods are replaced with walking legs. Image credit: Erin Jarvis
It was the first hint that in sand fleas, as in other animals, the combination of Hox proteins in a body segment might determine its identity. In a second study, this time led by former postdoctoral fellow Arnaud Martin, the Patel lab team sought to make sure that the correspondence between the Hox pattern and limb development wasn’t merely a coincidence. One at a time, the researchers functionally disabled the genes encoding each of the sand fleas’ Hox proteins—with dramatic results. Disabling a gene for a Hox protein that normally works in the abdomen caused walking legs to develop where swimming legs would normally be found, and the effects of disabling the genes for the other Hox proteins during development were no less striking. “It’s actually pretty easy to change between different types of limbs just by changing the developmental program with a single Hox gene,” Jarvis says. “Limb types can be switched around sort of like this modular Mr. Potato Head.”
The sand flea’s modularity isn’t an anomaly. It belongs to a group of organisms called arthropods, which includes insects, crustaceans and arachnids. Over the course of evolution, the arthropods’ simple segmentation scheme and many types of similar appendages—used for feeding, walking, flying, sensing, swimming and more—have made them extremely adaptable. This developmental flexibility, says Jarvis, may be one of the reasons arthropods are arguably one of the most successful groups of organisms on Earth.
The Hox hunt
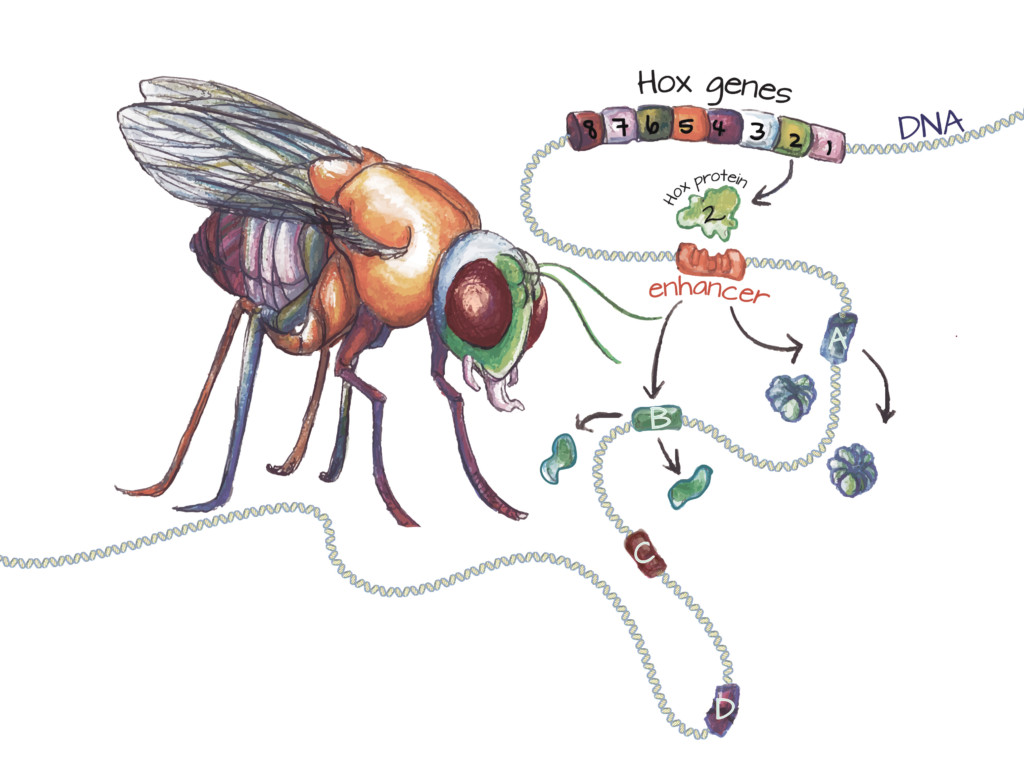 Hox genes encode Hox proteins: In all animals, the development of each body segment is orchestrated by specific Hox proteins. In the fruit fly, the second Hox gene (green cylinder) encodes a Hox protein responsible for patterning part of the head. The Hox protein binds to specific enhancers (slotted orange cylinder). After this binding event, the gene or genes this enhancer controls (A and B) are activated. Genes lacking this enhancer (C and D) are not activated by this Hox protein. Once a gene is activated, the cell produces the protein it encodes. Since Hox protein 2 is active in the head, gene A may encode a protein that promotes development of the mouth parts, while gene B may encode a protein that suppresses the development of legs. Image credit: Ashley Truxal
Hox genes encode Hox proteins: In all animals, the development of each body segment is orchestrated by specific Hox proteins. In the fruit fly, the second Hox gene (green cylinder) encodes a Hox protein responsible for patterning part of the head. The Hox protein binds to specific enhancers (slotted orange cylinder). After this binding event, the gene or genes this enhancer controls (A and B) are activated. Genes lacking this enhancer (C and D) are not activated by this Hox protein. Once a gene is activated, the cell produces the protein it encodes. Since Hox protein 2 is active in the head, gene A may encode a protein that promotes development of the mouth parts, while gene B may encode a protein that suppresses the development of legs. Image credit: Ashley Truxal
Far from being a mere curiosity of P. hawaiensis, the Patel lab’s results square with mechanisms found in other arthropods, and the same general process is found in almost all animals. Even subtle changes in the way Hox genes are used can drive body plan evolution—for instance, limbs can be made a little longer or shorter. And according to Jacques Bothma, a postdoctoral fellow researching animal development in Professor Hernan Garcia’s lab in the Department of Molecular and Cell Biology at UC Berkeley, repurposing or modifying a useful genetic program, such as the instructions for making a limb, is markedly more efficient than evolving a whole new program from scratch.
While Hox proteins have accelerated the pace of animal evolution, discovering all of them proved to be a slow process. In 1978, when Professor Edward Lewis reported his Nobel Prize-winning work on the development of the fruit fly Drosophila melanogaster, he hypothesized that there would be several Hox genes hidden in the genome. According to Bothma, “there were all of these groups who were looking for [the undiscovered Hox genes] all over the world because they knew they were important from genetic work.” But no one had been able to hunt down the other genes.
Close to capturing the remaining Hox genes were Michael Levine, Ernst Hafen, and William McGinnis, then members of Professor Walter Gehring’s lab at the Biozentrum of the University of Basel in Switzerland. In 1983, the researchers—all of whom are now professors—found a clue in Lewis’s manuscripts. Lewis had hypothesized that all modern Hox genes arose from a single ancestral Hox gene, repeatedly duplicated and mutated into multiple forms. Thus, the researchers hypothesized that all Hox genes should have some DNA sequence in common.
Suspecting they were on to something, the researchers used parts of a known Hox gene as a probe, searching the fly genome for similar sequences. And finally, the Hox genes succumbed to the chase. Bothma, who completed his PhD in Levine’s lab in the Department of Molecular and Cell Biology at UC Berkeley, recalls Levine’s account of the discovery. “They found the key to getting Hox genes,” Bothma says, “and then they got all of them at once.”
The DNA sequence shared by the Hox genes is especially important. It encodes what the researchers named a homeodomain, a stretch of 60 amino acids that’s one of the defining features of Hox proteins. The homeodomain is what makes each Hox protein a transcription factor, endowing it with the ability to bind to specific enhancers that activate developmental genes. Once the DNA sequence encoding the homeodomain was known, researchers began to find Hox variants in all animals—mice, worms, even humans. According to Bothma, “people before had this sense that there would be mouse genes involved in developing mice and there would be people genes involved in developing people.” But the discoveries of related Hox genes in more and more animals dissolved that notion completely.
In the years that followed, there was an avalanche of discoveries about animals’ shared developmental pathways. Researchers in Professor Gregor Eichele’s lab at the Baylor College of Medicine and Professor Thomas Kaufman’s lab at Indiana University found that replacing one of a fly’s Hox genes with the chicken variant of the gene doesn’t hinder the fly’s development, despite the fact that the last common ancestor of flies and chickens lived 673 million years ago. Beyond the striking similarities of the Hox genes themselves, many animals regulate the Hox genes in analogous ways.
As Lewis predicted, in several species the Hox genes cluster near each other on a chromosome. The position of each Hox gene in the cluster has special significance: in many animals, the order directly corresponds to the order in which body segments develop. The first gene in the Hox cluster encodes the Hox protein that is the first to act. This first Hox protein turns on genes that specify the characteristics of the first segment to develop, while the last gene in the cluster encodes the Hox protein that works last, activating genes involved in posterior development. Jarvis and her colleagues in the Patel lab have discovered that this property of Hox genes, known as colinearity, exists in some form in the sand flea—just as it does in distantly related animals.
All these similarities are, seemingly paradoxically, one of the keys to animals’ adaptability. Each Hox protein only specifies the identity of one segment in an animal, telling a group of cells that they are part of the abdomen, for example. The characteristics of that segment—for instance, whether it should grow wings or legs—are actually controlled by the genes the Hox protein activates. Since each Hox protein determines which genes to activate based on whether the genes possess an enhancer sequence just six DNA bases long, altering the characteristics of any body segment is always just a few mutations away.
Further exemplifying the flexibility of the conserved Hox program, some animals have done away with colinearity altogether. In a recent study, David Weisblat’s and Dan Rokhsar’s research groups at UC Berkeley collaborated to sequence and interpret the genome of the leech Helobdella robusta. In this leech, the Hox genes are dispersed across the genome. “The Hox locus is just shattered,” Bothma says. “All the Hox genes are just all over the place. The leech—it’s a degenerate, a fallen angel.” But perplexingly, in spite of the complete loss of colinearity, the Hox genes seem to function properly in the beautifully segmented leech.
A game of telephone
Perhaps unexpectedly, the leech isn’t really an outlier. Despite the strict adherence to colinearity in some animals, the Hox cluster is known to be reorganized or even scattered across the genome in a handful of other animals. These animals must have developed other ways to choose which Hox proteins rule each segment of the body, but for now they largely remain unknown.
When it comes to understanding the multitude of ways animals have employed this developmental machinery during evolution, maybe it’s naïve to expect a single unifying principle. The same could be said for multicellularity. It has evolved, as far as we know, over a dozen distinct times in the history of life. Some mechanisms of multicellularity are similar to one another; some are completely different.
These complexities are a natural consequence of the molecular mechanisms of evolution. Mutations, mostly random, are the seeds of life’s diversity. Slight modifications to genomes accumulate over the protracted timescale of evolution. Sometimes, a chance mutation results in a major evolutionary leap. Rarely, a truly original sequence is composed. Each of these incremental molecular changes may have consequences for the whole organism’s fitness—for better or worse. A genome’s evolutionary success, defined by whether it is passed on to future generations, depends on how well the culmination of billions of years of mutations have adapted it to its environment.
Although the imprints left by evolution are evident in every organism, it’s not always easy to observe evolution in progress. It is, however, simple to simulate. If you liked this article, make a few copies of it. In each of these offspring, create some mutations. Move a sentence or cut out a whole paragraph. Scramble a few words or insert a new phrase. Then place each new article in a new environment: pass it on to someone else.
These mutations may improve the article or leave it relatively unaffected. They might damage the article, making it less likely that the next reader will pass it on. The modifications you make to the text will, in part, determine its fitness in the hands of the next reader. Time will reveal whether these changes were successful. As in evolution, each new iteration of the article will diverge further and further from the original composition—if it survives.
Cover image credit: Ashley Truxal
This article is part of the Spring 2016 issue.



