One fish, two fish, rockfish, new fish
Reeling in the mysteries of aging

Our Fishy Neighbors
Off the coast of California exists one of the most diverse animals in the world: the rockfish. Over 150 different species of rockfish have evolved in the last 10 million years, an explosive burst of diversity rarely seen in nature. For UC Berkeley Professor of Integrative Biology Peter Sudmant, this biodiversity is a treasure trove of possibilities for research in his lab. Not only do the number of unique rockfish make them a great model for studying evolution, but they are also useful in studying longevity thanks to their range in lifespans—from 11 to over 200 years.
Although rockfish have never traditionally been used in research labs, the Sudmant lab is leading efforts to develop tools to study them. Their recently published study and ongoing projects have resulted in several new discoveries in aging, evolution, and species diversity, thanks to rockfish’s special attributes. “I was really enthralled by the diversity of lifespan among organisms on earth,” Sudmant explains. “I wanted to find a set of species that were closely related, and yet had differences in lifespan. I almost couldn’t believe how extraordinary rockfish were when I first learned about them.”
Methuselah's Zoo: Exceptionally Long-Lived Animals
Stories about longevity appear in almost all cultures and religions. Professor Steven N. Austad at the University of Alabama, Birmingham, wrote a paper in 2009, refocusing the scientific interest in human longevity—and coining the phrase “Methuselah’s Zoo.” Methuselah is one of the oldest Biblical characters, living for nearly 970 years. Rockfish may not have lifespans as long as Methuselah, but they have achieved lifespans that alchemists from long ago only hoped. This fact, combined with rockfish’s variance in lifespan, makes them one of the most important animals in Methuselah’s Zoo.
Nature contains a remarkable diversity of lifespans. The mayfly has an adult lifespan of only 24 hours, whereas the Greenland shark can live for over 300 years. How long an animal can live is typically explained through its unique biology and needs. To better understand the longevity of a species, scientists often assay an organism’s life history—the patterns of survival and reproduction events during the life of an organism. For example, longevity has a strong correlation with body size. The hypothesis is that larger animals live longer than smaller ones. For many years, lifespan was said to be the inverse of metabolic rate, or the number of calories an organism burns as they perform basic life-sustaining functions. Larger animals tend to do everything more slowly. Since they do not have to worry about fleeing from as many predators, they often move more slowly and are pregnant for longer. Adding to this trend, larger animals tend to age more slowly, too.
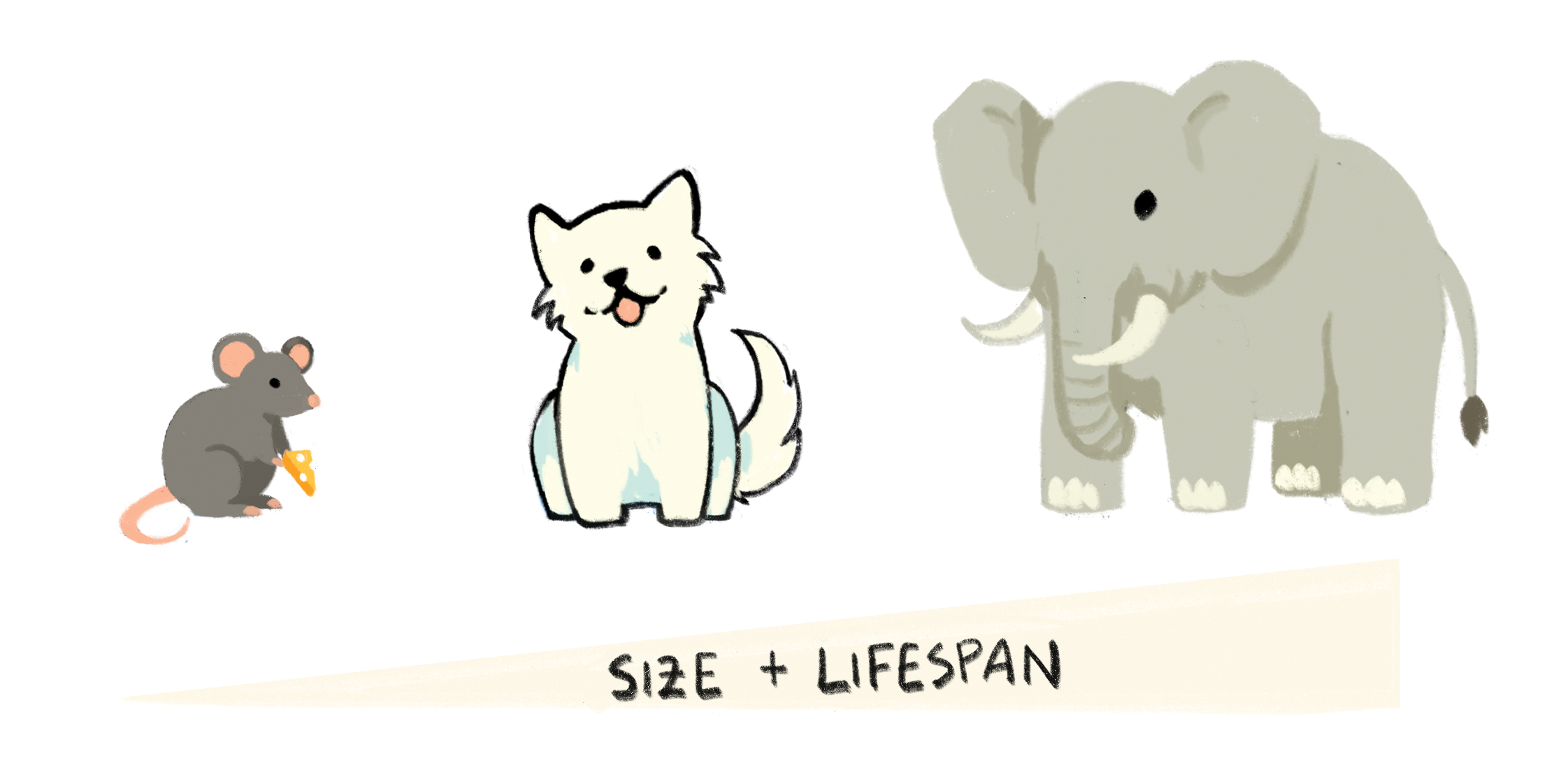
Of course, any natural trend is created to be broken. Rockfish stand out as a potential exception to the trend, since they can live for almost 200 years despite their relatively small size, especially when compared to larger species like sharks and whales. While their unusual size-to-lifespan ratio may be partly explained by the environment they live in—the longest-lived rockfish spend their lives in frigid, deep waters where their metabolism is extremely slow—this exception is still remarkable and studying why and how rockfish age could answer several questions about aging.
The Newest Non-Model Organism on the Block
Although rockfish may be the perfect species to understand aging and evolution, most scientists have not yet developed the necessary techniques to best study them. Instead, researchers are more likely to use model organisms, non-human species that they raise in the lab, to better understand biological processes. Typically, model organisms are easy to maintain and breed in laboratory settings, often by having short generation times, small body sizes, and short lifespans. Current examples include mice, fruit flies, nematodes, and yeast. These organisms are synonymous with science—they are the species names that repeatedly come up in textbooks, papers, and pop culture.
Working with model organisms affords scientists more protocols, infrastructure, and resources to rely upon. “When I was working in a yeast lab,” explains Department of Molecular and Cell Biology graduate student Sophia Adler, “I could Google protocols and find 20 to 30 papers with different examples and conditions, which was super helpful. If I try to do that in [a non-model organism], I might get one paper—but also maybe none at all—and that happens all the time.”
Studying non-model organisms such as rockfish requires overcoming many more hurdles. While entire journals are dedicated to experiments in model organisms such as yeast and mice, only a handful of articles have been published about rockfish. When researchers work with non-model systems, one of the biggest challenges they must overcome is developing and fine-tuning new protocols. Although many methods can be adapted from other systems, much of the foundation must be laid from scratch.
Luckily, rockfish check many boxes that might also make them easier to study than most other non-model organisms. Many species of rockfish can be brought into the lab space and survive in aquariums. It is also relatively easy to gain access to them through commercial fisheries and farming, oftentimes without having to kill the fish in the process. Additionally, UC Berkeley is uniquely situated very close to the Gulf of the Farallones, located just outside the Golden Gate Bridge, where dozens of rockfish species live.
“We are very lucky that many species of rockfish live off the California coast,” says Sudmant. “A graduate student in my department, Alexander Stubbs, is a commercial fisherman, and he had a boat. So, we went fishing! We also enlisted the help of other fishermen, the National Oceanographic and Atmospheric Association (NOAA), and a number of museums.”
Sudmant’s collection of rockfish samples also implies that studying non-model organisms is no longer as difficult as it used to be. “People use worms and yeast to study lifespan, because they are wonderful organisms you can grow in the lab,” Sudmant says. “You can even do that with mice—even though they live for two years. But you can’t do that with rockfish because their lifespan is longer than a researcher, let alone a graduate student. Rockfish are never going to be a model organism in that sense.”
“But the world is changing now,” Sudmant continues. “Technologies allow us to completely deconstruct the genomes of all kinds of organisms. Simultaneously, technological advances in the lab are allowing us to make cellular models for the first time, and do all kinds of very cool experiments.”
So, while rockfish may never have the traditional and vaunted status of model organism in the old sense of the word, they have become one of the first species to herald a new age of biological science research. In this innovation, the Sudmant lab leads the charge in developing the first protocols and techniques to determine how to best study rockfish and, in a larger sense, non-model species as a whole.
Going Fishing on a Scooter
Department of Integrative Biology graduate student, Stacy Li, is one of the first researchers to take on the challenging and oftentimes daunting task of developing the proper protocols and techniques necessary to prepare rockfish for specific laboratory experiments.
Earlier this year, the Sudmant lab was receiving samples of “fin clips,” small hole-punches of fish fins that are a relatively non-invasive way of collecting DNA samples. Based on technical application notes about DNA extraction, Li believed that they would be able to collect high molecular weight DNA. This type of DNA is intact and contains long reads of sequences. High molecular weight DNA is of particular importance as it has haplotype information. Haplotypes are blocks of DNA which are typically inherited together, but are sometimes broken up over many generations, resulting in genetic variation. By extracting high molecular weight DNA, researchers may be able use haplotype information to understand how rockfish evolved.
But Li was not seeing these haplotypes in the fin clips they were processing. There was no DNA present in their samples, let alone the long DNA they’d need for computational analysis. “If the problem was the biological sample,” explains Li, “I didn’t want to waste any more of the precious ones we’ve been sent—I was just going to have to find some more fish fins.” Collaborator samples were limited, and Li was keen to minimize the number of samples that needed to be collected due to the significant effort involved.
So, Li hopped onto their trusty scooter, took their bag and some cash, and went over to Monterey Fish Market. Through the store’s Instagram page, Li had noticed that they sold several varieties of rockfish. At 9 a.m. on a weekday morning, Li went up to the fishmonger who was laying out some rockfish on ice.

“My first question to him was: ‘How long has it been since this fish died?’” says Li. “Time from death and temperature storage are two of the biggest contributors to determining whether or not we’d be getting high molecular weight DNA, but he didn’t know the exact answer—guessing that it could be anywhere between one and three days.”
While this number is excellent for human consumption, it wasn't the best condition for DNA. This experience confirmed Li’s fears: even exemplary standards for fish storage for human consumption were not ideal conditions to study biology. Regardless, Li wanted to give it a shot and scooted back to campus with their bounty.
Back on campus, Li made careful notes and decided to collect samples from their fish’s organs. They planned out storage conditions and cleaned their fish for processing. Generally, the fish fin, heart, blood, spleen and gonads should contain the most amount of high molecular weight DNA.
“As I’m clipping away at this fish with a picture of fish anatomy as a reference, my whole lab was standing around me guessing what the organs were,” Li laughs. “It was a fun and weird thing. All the biology I’d done before had been in petri dishes and little tubes.”
Several hours later, Li was left with tubes of samples containing high molecular weight DNA, and the body of their mutilated fish—which they proceeded to take home for dinner.
Understanding the Evolution of Aging
What Li successfully obtained from their rockfish samples lay the framework for the Sudmant lab to answer several questions about the evolution of aging. Rockfish are powerful models to study evolutionary relationships because they make up a clade, or group of organisms believed to have evolved from a common ancestor, for over 10 million years. This large amount of time means that individual species had more time to accumulate mutations, while conserving bits and pieces from a common ancestor. Mutations change the structure of genes, and usually occur at a steady rate. The more species that exist means that more pieces of the common ancestor have been conserved. Thus, by having more species that evolved from the same ancestor, researchers have more data to put together the original genome of the original ancestor. In the process, they can also figure out what genetic changes underlie the presentation of certain traits in the organisms of today.
When assembling genomes, scientists often rely on a high-quality reference genome. Making a reference genome is a complex process. In an ideal world, the entire length of the genome would be read by a machine, outputting the different nucleotides of the genome sequence (A, T, C, and G) as it goes. However, because this technology doesn’t exist yet, scientists must assemble the genome from small fragments of DNA.
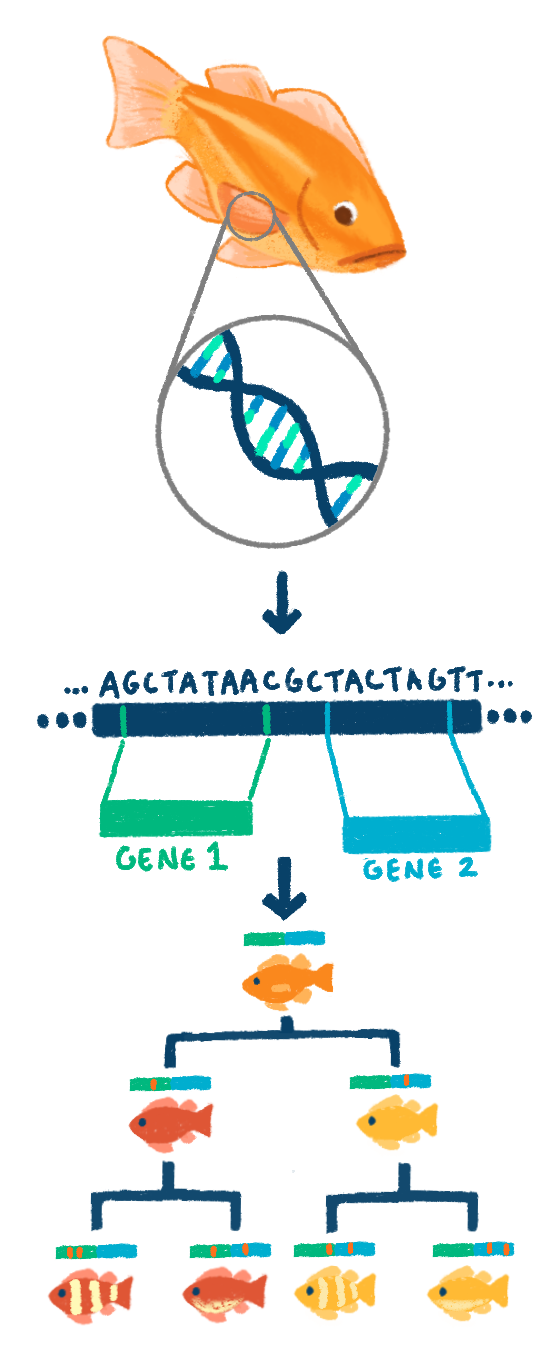
This process is akin to taking 10 copies of a newspaper and shredding it. Some of the pieces will overlap, allowing you to rebuild the pages. If a particular page or fragment has seven copies of the same words at the same position, the “read depth” is said to be 7x. Parts of the rebuilt newspaper may have only one or two copies, and would therefore have a lower read depth. A lower read depth gives scientists less confidence that they truly have the correct sequence of words and that a typo was not introduced—but many times that’s all they have to work with. Ultimately, by chopping many copies of the genome into fragments, researchers can then sequence those fragments, called “reads”. By noting where the reads overlap, scientists can put the DNA fragments in the right order.
Previously, reads were around 1,000 nucleotides long, which made some parts of the genome, especially where there were many repeats, unclear. However, the Sudmant lab was able to use recent long-read sequencing technologies, which increased the length of the reads to up to 100 thousand nucleotides. Longer reads allow for more clarity. In other words, each individual shred of the newspaper is longer, so instead of using just letters to put the page back together, you are using sentences, phrases, and paragraphs. When it comes to rockfish genomes, not only do long reads make for a more accurate assembly, but they also allow for more confidence. By using this method, the Sudmant lab was able to build high-quality reference genomes for many of their rockfish.
These long-read sequences allow scientists to faithfully reconstruct the evolutionary history of every gene in the rockfish. With this capability comes an incredible power to detect any genetic changes associated with the longevity of long-lived rockfish relative to short-lived rockfish. In fact, researchers in the Sudmant lab worked with 88 different rockfish species to pinpoint 137 genes that were correlated with an increased lifespan. However, lifespan in rockfish is strongly correlated with both body size and environmental factors. What this finding means is that some of the genes associated with lifespan may primarily act by influencing growth or may facilitate adaptations to cold, deep-ocean environments where long-lived rockfish are more likely to live, rather than directly promoting longevity.
To identify genes associated primarily with longevity, the group had to parse out which genes were associated with lifespan independently of body size and environment. The group used a statistical model to identify 56 genes that were independent of rockfish body size and the ocean depth at which they were found. These included several genes that were known to increase lifespans across other animals, including insulin and glucose signaling and butyrophilins, genes which regulate the rockfish immune system and are known to suppress inflammation in aging humans.
From Fish to Humans: What Would Translation Look Like?
“We found that butyrophilins have a higher ‘copy number’ in ultra-long-lived species,” Sudmant explains. “This highlights a specific set of genes and pathways that might be important to follow up in humans.”
Insulin signaling, identified as another one of the primary genes related to longevity in rockfish, is one of the most important pathways in aging. Rockfish also have their own insulin-like factors which have undergone different changes due to mutation. Speculatively, scientists could use an insulin-like factor in long-lived rockfish and observe changes when inserted into the genome of short-lived rockfish. If the new gene indeed extends the fish’s lifespan, then these factors could be inserted into genomes of mammals more closely related to humans than fish. Extensions in lifespan in model organisms may suggest that these factors could be used in humans next.
However, the lead author on the study, postdoctoral researcher Dr. Rohit Kolora, is cautious when talking about the translation of longevity in rockfish to humans. “Longevity from rockfish is not something we can directly translate into humans,” Kolora explains. “Something that increases longevity in deep-sea fish may not work in freshwater fish. What works in freshwater fish might not work in birds. And what works in birds might not work in humans.”
But Kolora still has big plans for rockfish and the problems they might solve some day. From better sequencing reads to molecular manipulation, Kolora is hopeful that better genetic datasets and tools will keep rockfish studies moving forward. “We have the capability to culture rockfish cells, and manipulate them with genomic tools to remove specific genes from the rockfish and see how the removal impacts the health of cells,” Kolora says. “There’s a lot more to learn when it comes to rockfish longevity.”
Besides lifespan, translational studies from rockfish would also provide insight into health span—or how long an individual stays healthy. One of the criticisms of current human aging research is that there is little use to living for a long time if it means being tied to machines, medications, and end-of-life care for years or even decades. Because rockfish are wild animals, who spend their 200 years eating and hunting, they must maintain fitness into old age.
What would health span studies look like in humans? Studying longevity isn’t feasible because such research would require a clinical trial in which a participant may outlive the clinician. Instead, researchers may look at certain diseases which arise only in old age, such as Parkinson’s or Alzheimer’s disease. They could then see if a cohort treated with a drug mimicking a rockfish biomolecule suffers from fewer cases of the disease.
While it may be years before rockfish studies are translated into humans, we may see these genes manipulated in mammals, and perhaps even our pets, sooner than that. “Aging is a disease that can be solved,” says Dr. Juan Manuel Vazquez, a postdoctoral researcher in the Sudmant lab. “It is no longer an intractable problem.”
Rockfish Through History – From Bountiful to Collapse
Even besides aging and evolution, rockfish can answer other questions, such as the role humans play on rockfish ecology, and on the environment as a whole. A staple fixture in the Sudmant lab is the fish freezer (nicknamed “British Columbia” as a nod to Sudmant’s Canadian roots), filled with dozens of samples of rockfish collected over several fishing expeditions. One of the frozen residents is the boccacio. This species of rockfish was once prevalent off the California Coast, and its large size lent itself for human consumption. Boccacio live for almost 50 years—much longer than most fish species that are farmed. However, because of their long lifespan, when the population was decimated in the early 2000s due to overfishing, it left the younger fish to repopulate. Because the boccacio are so long-lived, changes in their population structure have persisted a long time.
“This population collapse created a massive human-shaped hole in the boccacio population,” explains Li. “It’s important to study this phenomenon because California is flush with resources for human consumption and living—especially in the Bay Area. We obtain so much of our diet from local production—and that includes rockfish. If we are not careful about the way we conduct stewardship of our natural resources, we could end up decimating more populations.”
Boccacio fisheries have been recovering since 2004 due to careful management from California Fish and Wildlife. But in this situation, rockfish present an interesting study of a human-made phenomenon that impacts a species that lives for an unusually long time. And most importantly, their ecology is tightly entangled with ours.
Modern examination of rockfish is one way the boccacio population structure is being studied. The other method is to examine historical rockfish to understand the human impact on rockfish over time. This kind of study can only be done longitudinally—a research design that involves repeated observations of the same variables over long periods of time—and the Sudmant lab is one of the few researcher groups able to lead a project due to their extensive groundwork on how to process rockfish samples in the laboratory.
In a collaboration with NOAA and Dr. Milton Love, a research biologist at the Marine Science Institute at University of California, Santa Barbara, the Sudmant lab obtained boccacio samples over the course of 15 years—from 2004, when boccacio fishing became regulated by the federal government, to 2019. The lab received 400 samples.
The clips, about a hole-punched millimeter in size, were used to get short fragments of DNA, which were compared to a reference genome. A representative subset of these samples would allow the researchers to see how the immense depletion in 2004 to 2019 would affect the genetics of a population.
“This isn’t just a story about fish,” Li elaborates. “It’s also a story about how humans are affecting the world around them. If we are not careful, we could end up creating population changes and affecting the genetics and genomics of the population for generations to come.”
Just keep swimming: what’s next for the rockfish?
“The thing that I find most exciting about our study is that we identify both the causes and the consequences of extreme lifespan,” describes Sudmant. “It means that not only are there genetic changes that allow these fish to live so long, but, because they live so long, long life is also reshaping patterns of genetic diversity.”
There is still more work to be done. Statistical methods allowing researchers to look back in time have shown that there is a very tight linkage between size of a population and lifespan. That is, longer lived species have smaller population sizes than shorter ones.
“We are continuing to study the genetic diversity of these fishes, particularly at the population level,” Sudmant explains. “This project was more about comparing between species, not within species and I am really excited to understand the relationship between the extraordinary lifespan of these fishes and their rapid speciation.”
The lab also plans to strengthen their understanding of the functional consequences of some of the genes they found, by growing fish cells in the lab and manipulating them. Similarly, there are also efforts to build more long-read reference genomes. “If we have better reference genomes,” Kolora explains, “we may be able to make stronger and better conclusions about the evolution of aging.”
In the meantime, the Sudmant lab is already revolutionizing our understanding of aging and evolution. “Aging is as fundamental a biological trait as DNA is part of cells,” explains Vazquez. “It is a programmed process. Rockfish are simultaneously helping us understand one of the most fundamental processes of life, but also helping us understand how that process came to be.”
------- Samvardhini Sridharan is a graduate student in molecular and cell biology
Design by Liya Oster
This article is part of the Spring 2022 issue.