Let’s dive into our genomes. Each of us carries 2 meters of DNA in every cell, which in turn carries the information that makes us all unique. However, some regions in the genome are extremely tricky to navigate and decipher. These regions contain many genetic repeats, which confuse current DNA sequence analysis technologies, making them difficult to study. The centromere is a particularly troublesome region, but scientists at UC Berkeley have discovered a clever phenomenon to help crack this DNA code.
The centromere, which is the cinched-in, central region of DNA on every human chromosome, is essential to maintaining genomic information. Cells divide throughout our lifetimes in order to grow and maintain organs and tissues, and each new cell must contain an intact, whole version of the genome. During cell division, the centromere serves as an anchor point to pull each chromosome into the newly formed cell. Loss of centromeric function results in chromosomes being incorrectly lost or gained, which affects aging, cancer formation, and tissue development.
 Recombination, which mixes up maternal and paternal DNA, is less likely to occur in centromeric regions.
Recombination, which mixes up maternal and paternal DNA, is less likely to occur in centromeric regions.
In addition to this key role in cell division, the centromeres prevent DNA from getting reshuffled and exchanged as our genes pass through generations. These preserved DNA sequences have the potential to provide insight into the evolution of human centromeres.
Now, a new study in eLife by Dr. Sasha Langley and colleagues at UC Berkeley, Lawrence Berkeley National Laboratory, UC Santa Cruz, and UC Davis shows that all human chromosomes have very large regions of DNA at and around the centromere that are always inherited together. They call these regions cenhaps, or centromeric haplotypes. A single chromosome can have several cenhaps of different sizes along the entire centromeric region. Cenhaps can be used to identify differences in centromeric DNA sequences between humans and indicate functional roles in key cellular processes, such as cell division.
The discovery of cenhaps has not only opened the door to investigate many unanswered questions about centromere function, it has also exposed fascinating insights into our evolutionary history. Many of these cenhaps have ancient lineages that are more than 600,000 years old. According to Langley, "different chromosomes seem to have different histories and different levels of diversities [in their cenhaps], thus making each chromosome's sequence unique.
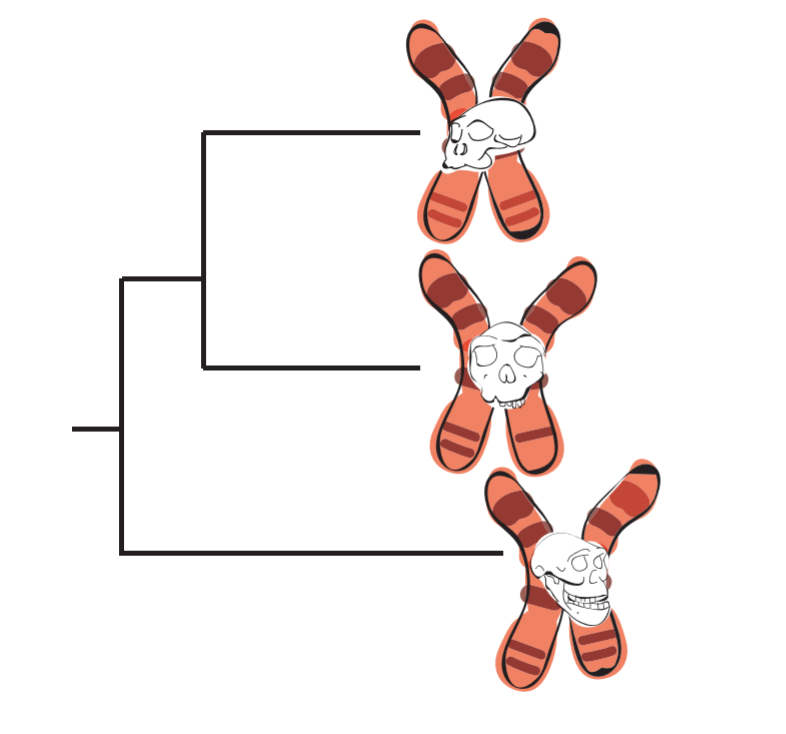 Different centromere variants, or "cenhaps", can act as "genetic fossils" that inform us about our evolutionary history as humans.
Different centromere variants, or "cenhaps", can act as "genetic fossils" that inform us about our evolutionary history as humans.
In some cases, the ancient cenhaps allow the researchers to speculate on the origins of associated genes. For example, Langley identified two cenhaps with very different characteristics on the same chromosome. When comparing these cenhaps across genomes, she found that non-African genomes contain highly distinct sequences with primarily Neanderthal DNA, while African genome cenhaps revealed a less clear archaic lineage. When she examined the non-African haplotypes, she found 34 genes for odorant receptors, perhaps suggesting a link to smell and taste determinants in Neanderthals. Odorant receptors “tend to be in those regions in clusters, so it wasn’t super surprising,” Langley explains, “but it will be interesting if people start to look at the functional consequences of them all travelling together."
Langley found that another chromosome also has two cenhaps, one of which had a high affinity to Neanderthal DNA. However, the second haplotype has DNA inherited from a human species even older than Neanderthals or Denisovans. Other human chromosomes showed older lineages as well, suggesting that the lack of rearrangement at centromeric regions with each generation has allowed archaic DNA to persist in our centromeres to the present day.
The researchers are particularly interested in addressing fundamental scientific questions that connect the cellular functions of centromeres to the sequences found in cenhaps. Langley says that this is a “big step forward in that [cenhaps] really give us a way to ask the functional questions that people haven’t been able to address before.” The human genome is now more accessible for all scientists, as cenhaps provide a means to explore the tangled and tricky centromere and investigate evolutionary variation.
Maiko Kitaoka is a graduate student in molecular and cell biology
Design: Kirsten Isabel Verster
This article is part of the Fall 2019 issue.