Taking evolution by swarm
Flying toward evolutionary answers

One in a trillion: metrics of biodiversity through the lens of speciation and hybridization
The diversity of life has always shocked naturalists and evolutionary biologists. During Darwin’s celebrated voyage, he emphatically noted how dissimilar the birds from different islands, only a few miles away from one another, seemed to be. With contributions from famous ornithologist John Gould, Darwin described many new species—formal classification for groups of organisms that are similar, but capable of exchanging genes or interbreeding to produce viable and fertile offspring. These insights catapulted Darwin to fame a few short months later, when he described the idea of evolution by means of natural selection based on these observations.
Centuries later, research has still only scratched the surface about what the vast number of species tells us about the world around us. Scientists estimate that there are between 5.3 million and 1 trillion different species on our planet. The margin of error on this estimate can be described as immense. Keeping track of the number of species on Earth is more than just a scientific pastime, it’s also an important metric to monitor ever-changing biodiversity. Discovering and cataloging new species allows researchers to create more detailed studies of population diversity. Simultaneously, many of the planet’s species are quickly going extinct. Currently, our planet is in a human-caused mass-extinction phase, with many species disappearing almost as quickly as they are discovered. However, populations can also change by appearance of novel species, known as speciation.
Sometimes, speciation occurs due to geographic separation as Darwin observed in the Galapagos, but occasionally, groups from the same ancestral population evolve into separate species with no geographical separation at all. When divergent species produce offspring together, their progeny are known as hybrids. Hybridization is an important genetic and evolutionary process and allows scientists to explore biodiversity in all its forms.
Hybrid animals present interesting models for studying evolution and biodiversity because they exist in a liminal space as offspring of animals that are different enough to separate but still similar enough to reproduce together. Mules are a famous hybrid between a horse and a donkey. Not only stronger than horses and donkeys, mules also tend to live longer—exceedingly important for owners who tend to employ them in intensive physical tasks. Despite these advantages, mules are sterile and unable to reproduce. Horse and donkey chromosomes are just too different to make fertile offspring, with horses having 64 chromosomes and donkeys having 62. Yet, horses and donkeys are still similar enough that they can mate. Understanding how and why hybrid offspring are different from their parents is an important field of study.
Dr. Doris Bachtrog, an integrative biology professor at UC Berkeley, is interested in speciation in Drosophilae, commonly known as fruit flies. Drosophilae are celebrated model organisms—quick and easy to breed, and, in the genomic age, cheap to sequence. There are hundreds, if not thousands, of species of Drosophila, and hybrids between them are common. In cases where hybrids are viable and fertile, there is genetic material introduced from one population of species to another, changing the gene pool. In a recently published study, UC Berkeley postdoctoral researcher Dr. Silu Wang and other Bachtrog lab researchers find that sex chromosomes—chromosomes involved in determining the sex of an organism—play an important and asymmetrical role in speciation. “I find speciation is a fascinating process that produces 8.7 million eukaryotic lives,” says Wang. “It is such a fascinating process to allow organisms to evolve from a single ancestral lineage to one with such great diversity.”
But Wang’s study does more than just tell us about speciation. It also brings up important questions about the changing methods in evolutionary biology research. A range of experimental and computational methods shine new light on rooted study systems and presents a promising foundation to study the natural systems around us.
The great fruit fly chromosome abnormality
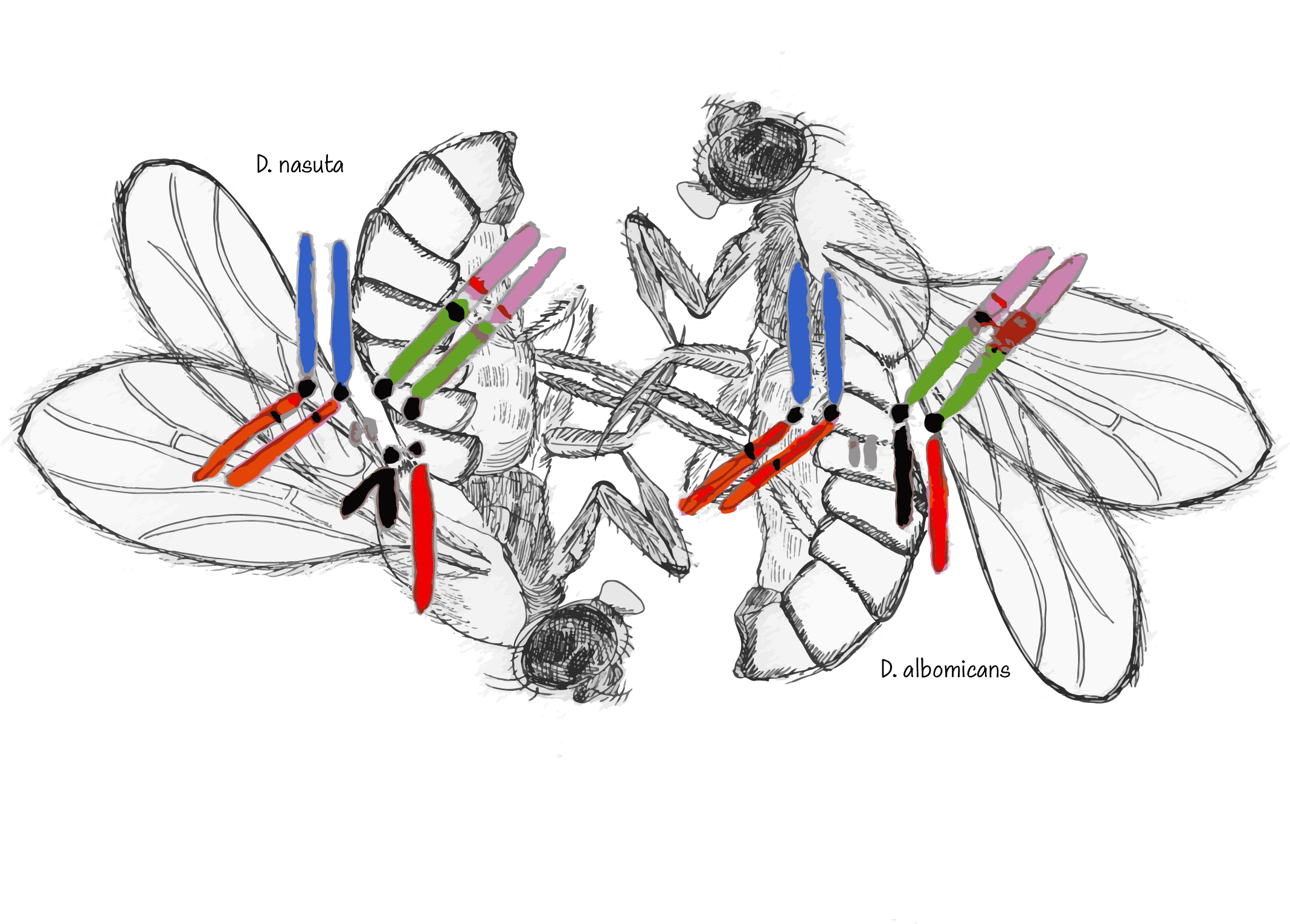
Where do most genetic incompatibilities manifest? To answer this question, the Bachtrog lab established a study system between Drosophila albomicans and its sister species Drosophila nasuta. D. albomicans and D. nasuta are morphologically indistinguishable, but are found in different places in the world. While the two species formed due to geographic separation, synthetic hybrid populations provide a valuable system to study patterns of genetic ancestry. Importantly, hybrids show little incompatibility between genes in their offspring, allowing the scientists to specifically identify when genetic material is transferred and its source.
Compatibility within hybrids is particularly significant as D. nasuta and D. albomicans have unique sex chromosome structures. D. nasuta shows the same chromosome structure of most Drosophila species. Meanwhile, D. albomicans has what is known as a neo-sex chromosome pair in which different parts of the chromosomes have fused together. The fact that these very different chromosomal structures can come together to produce viable offspring provides a unique model to study sex-chromosome evolution.
A long-standing theory is that the inability of two species to successfully breed is disproportionately sex-chromosome linked. Researchers were keen to determine how sex chromosomes affect the patterns of genetic material transferred between two species, astutely looking for biases in where genetic material was being transferred, and bias they did see.
Genetic incompatibilities on the sex chromosomes are described as snowballing: as genomic differences accumulate over time, there is a faster and faster rate of differences in chromosomal structure. That is, the more generations of flies are raised, the more genetically distinct the progeny are from their parents. But there is one important caveat. Studies like Wang’s can test whether the snowball effect occurs only under well-defined and relatively controlled experimental circumstances. The reality of the snowball effect in the “real world” is still something many scientists are interested in learning more about.
It's all relative—ancestry patterns in a hybrid swarm
The Bachtrog lab observed patterns of genomic differentiation over more than 62 generations of hybridizations and hundreds of flies, known as a “hybrid swarm.” Hybrid swarms are particularly useful in determining which parts of the sex-chromosome are inherited from each parent, relative to the rest of the genome. Wang analyzed these metrics by looking at genomic fragments. If the D. albomicans and D. nasuta genomes contribute equally to inheritance, the proportion of ancestry should be 50:50 and randomly distributed. If one species contributes more to a region than another, most flies would contain genes originating from that species.
Wang was able to extract a lot of information from the haplotypes of her hybrid flies. Haplotypes are blocks of DNA which are typically inherited together, but are sometimes broken up over many generations, resulting in genetic variation. Over multiple generations, Wang noticed that a long stretch of one chromosomal region did not have ancestry turnovers—regions which changed ancestry from D. albomicans to D. nasuta, or vice versa—while the rest of the genome seemed to have relatively the same proportion of turnovers.
The result was unsurprising because of existing work on chromosomal structure and diversity, but became evidence to the importance of neo-sex chromosome evolution in hybridization research. Wang and her team probed further lines of inquiry, working hard to pinpoint exactly what mechanisms and biological processes were contributing to regions of low turnover.
Tokyo drifting toward higher gene frequencies
At the core of Wang’s research is the idea of intragenomic conflict—why some genes are preferentially inherited from one parent versus the other. While researchers can observe regional preferences of inheritance, another factor that may lead to these patterns are meiotic drivers, which are genes that may be harmful to the individual, but still somehow increase in frequency within the population. These genes are insidious, breaking the fundamental rules of inheritance where both the biological mother and father are equal as the meiotic driver spreads despite potential harm to its hosts.
Wang was particularly interested in how meiotic drive would affect the evolution of the neo-sex chromosome, especially given the context of the ancestry turnovers. Was the unequal contribution from each parent due to “genetic fratricide?” The answer wasn’t nearly as cut and dry as she thought. “It’s still hand-wavy,” Wang explains. “We don’t know how this meiotic drive evolved. What we do know is that these species occur within the same geographical area and might have contact zones—and when these species hybridize there might be selection against the hybrids, preventing further mixing.”
On a larger scale, meiotic drive puts a wrench into the idea of hybrid vigor like in the earlier example of the mule, which is seen in many other plant and animal species. After all, how can you have hybrids if there’s a genetic mechanism that specifically prevents hybridization in the first place? Whether or not hybridization helps a population survive is largely dependent on whether the introduction of new species to the genetic pool can present novel genetic adaptations that can help the two populations survive. But in species that have diverged for a long time, such as in horses and donkeys or even D. albomicans and D. nasuta, the populations have accumulated a lot of genomic differences that can be incompatible with one another and affect fitness. “If there is a choice to mate within the same species, versus with a different species, if the individuals make the wrong decision, it could be detrimental—and there could be long-lasting effects in the genome for many generations,” says Wang.
In many ways, this delicate, incompatible balance emphasizes how little we know about the forces that drive speciation. “All these evolutionary forces have to work as a symphony,” Wang explains excitedly. “And the cool part is that our lab has generated parallel hybrid swarms—so it’s not just one particular experiment ending up in a weird way.”
In fact, the symphony held up to a double encore. Wang’s team was able to show similar phenomena in two other hybrid cages. Not only was the experiment repeatable, but it also pushed the limits of what scientists were able to discover about species boundaries. Previously, these asymmetric ancestry patterns were not shown empirically in experimental systems in the lab. “We were really able to dissect the forces at play in this complex sex-chromosome system,” Wang explains. “Now we can really see why the effect of sex-chromosome evolution can be so conflicting and complicated.”
Towards a computationally intensive future
Wang’s research used many computational methods in addition to traditional experimental techniques when it came to Drosophila hybridization and maintenance. The work from many researchers in the Bachtrog lab uses a mix of these methods—though, as a field evolutionary biology is tending towards more computationally heavy research.
For example, Wang’s ancestry analysis used software, AncestryHMM, created by Dr. Russ Corbett-Detig, now an associate professor at the University of California, Santa Cruz, when he was a postdoctoral researcher at UC Berkeley. One advantage of Ancestry HMM was that it could use low-coverage data from many individuals to determine their ancestry. Low-coverage data can often be problematic in genomic studies because there is not enough information to determine where a region of the genome comes from.
“If you do not have information about the whole genome, you can look for variation,” explains Dr. Manjusha Chintalapati, a postdoctoral researcher in the Moorjani lab in UC Berkeley’s Molecular and Cell Biology department, who studies human evolution. “There are markers in the genome called SNPs, which are fixed – defining these area of the genome as one lineage or another.” As most regions surrounding SNPs are not broken up in the genome shuffle, researchers are able to extrapolate that larger regions of the genome come from a particular ancestral haplotype. Ancestry HMM leverages these ideas to determine ancestry.
When it comes to the analysis, the availability of data is key. “It is becoming easier and easier to generate high-quality large-scale genomic datasets,” Wang details. “There needs to be thorough and efficient computational thinking to extract information from all of it.” Meanwhile, computational thinking comes easy to researchers like Dr. Corbett-Detig and Dr. Chintalapati who study evolution through a purely computational lens.
“To be successful and at the cutting edge, any field is going to require some amount of computer science,” Corbett-Detig hypothesizes. “That’s not to say it will subsume entire experimental fields—you still need to be a biologist—but the scale and pace of data generation is increasing all the time, and what that means is we will need to write more scalable algorithms and programs if we want to use all that data.”
“I think there is a surplus of data today, and there will be even more in the future,” explains Chintalapati, “but the direction of the field towards computation largely depends on the questions people are going to ask. There are model systems [like Wang’s] where we can capture sex-chromosome evolution as it’s happening, but that’s really not feasible in all systems, especially in humans, which I work with.”
It's increasingly clear is that evolutionary biology is no longer the observational field of yesteryears. A mix of methods, from the experimental to the computational are becoming progressively more important to the scientific storytelling coming out of the research world today: as scientists continue to add tools to their toolboxes, the future of the field is far less myopic than it used to be.
The study system divide: natural populations versus experimental ones
Over her career, Wang has studied fruit flies, North American warblers, livebearer fishes, and tinamous. Understanding how species split has been a long-standing research goal of hers and understanding the genomic tensions of various evolutionary forces continues to be a topic of interest and what she plans to study in her own lab someday.
The idea that some genes confer organismal advantages that promote their transmission is not new. In the 1970s evolutionary biologist Leigh Van Valen coined the evolutionary term “Red Queen hypothesis,” in which species must continually evolve to survive in the face of their enemies. Hybrid species pose an interesting approach to study this hypothesis, as they have a higher resistance to their biotic and abiotic pressures.

“While sex-chromosomes are an interesting area, there are other things I am interested in. … I find intragenomic conflicts in general are a fascinating area to think about the nascent species boundaries,” describes Wang.
But when it comes to future projects, Drosophila are not the only system Wang plans to analyze to answer these questions. “Drosophilae are very, very useful to understand genomic tensions but they aren’t my favorite system to study experimental evolution,” Wang details. “My favorite systems are natural populations. We are at the stage where we need to think about these processes in more complexity in natural populations and natural biodiversity. That’s the main area we are marching to as a field.”
Driving this belief is empirical evidence on extinction. Every hour we lose approximately three species, a scary rate of biological loss. Characterizing the diversity of these populations before we lose them is uniquely important. “The more urgent priority is bringing quality population genomic and evolutionary genomic understanding for wild populations,” Wang emphasizes. “Hopefully insight about species boundaries and adaptations and their processes will help us understand the past and the future of these populations and contribute to their conservation.”
However, Wang is quick to acknowledge that a combination of natural populations and experimental evolution may be what it takes to understand speciation and hybridization in the end, despite where popular interest may lie. Model systems like the Drosophilae are critical in understanding what scientists must look for. Be it sex chromosome evolution, reproductive boundaries, or even developing methods to ask better questions—the natural relies on the experimental as evolutionary biology writes its next chapter.
------- Samvardhini Sridharan is a graduate student in molecular and cell biology
Design by Chenyu Zhang
This article is part of the Fall 2022 issue.